| Environments | PYTHON :: EASI :: MODELER |
| Quick links | Description :: Parameters :: Parameter descriptions :: Details :: Examples :: Algorithm :: Acknowledgements :: Related |
| Back to top |
INSRAW2 creates a raw interferogram using coregistered reference and dependent data sets. INSRAW2 supports the following coregistration configurations:
All resampling must be performed by the INSCOREG module.
You can, as an option, filter the interferogram by using an adaptive Lee filter.
| Back to top |
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
| Name | Type | Caption | Length | Value range |
|---|---|---|---|---|
| FILREF* | str | Database reference file name | 1 - | |
| DBIC_REF | List[int] | Database reference input channels | 0 - | |
| FILI* | str | Database input file name | 1 - | |
| DBIC | List[int] | Database input channel list | 0 - | |
| FLSZ | List[int] | Size of filter | 0 - 1 | |
| FILO* | str | Database output file name | 1 - |
| Back to top |
FILREF
The name of the file to use as the reference data set. The file must contain single-look complex (SLC) data. The pixels of the dependent file must align one-to-one with the pixels of the reference file. FILREF can be either in the original slant-range projection, or resampled to the dependent file, or in the projection of a third (i. e. common) reference file.DBIC_REF
The list of channel numbers from the reference file.FILI
The name of the file to use as the dependent data set. The file must contain single-look complex (SLC) data. The pixels of the dependent file must align one-to-one with the pixels of the reference file. FILI can be either in the original slant-range projection, or resampled to the reference file, or in the projection of a third (i. e. common) reference file.DBIC
The list of channel numbers in the dependent file. The number of channels must match identically with the number in the reference file.FLSZ
The filter size, in pixels, of the adaptive Lee filter to apply. You specify the filter as n x n, where n is an odd integer greater than or equal to five.FILO
The name of the output file to which to write the raw interferograms. The file will contain the same number of pixels and lines as the reference file and the number of channels you specified. Nonoverlapping areas will be processed as NoData. The file name you specify must not already exist.| Back to top |
| Back to top |
Generate a nonfiltered, single-channel interferogram between the reference file (in its original projection) and the resampled dependent file (generated by INSCOREG) using the default values.
from pci.insraw2 import insraw2
filref = r"C:\MyProject\Reference_File.pix"
dbic_ref = []
fili = r"C:\MyProject\Resampled_File_1.pix"
dbic = []
flsz = []
filo = r"C:\MyProject\Interferogram_1.pix"
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
Generate a nonfiltered, single-channel interferogram using the resampled file (from the previous example) as the reference file and a third file already coregistered to the resampled file (by INSCOREG). Note: The input files "Resampled_File_1" and "Resampled_File_2" and the output files "Interferogram_1" and "Interferogram_2" are in the same slant range projection as the original reference file. This technique can be used to coregister multiple files to a common slant range projection.
from pci.insraw2 import insraw2
filref = r"C:\MyProject\Resampled_File_1.pix"
dbic_ref = []
fili = r"C:\MyProject\Resampled_File_2.pix"
dbic = []
flsz = []
filo = r"C:\MyProject\Interferogram_2.pix"
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
Generate a filtered interferogram using a seven-by-seven adaptive Lee filter.
from pci.insraw2 import insraw2
filref = r"C:\MyProject\Reference_File.pix"
dbic_ref = []
fili = r"C:\MyProject\Resampled_Dependent_File.pix"
dbic = []
flsz = [7]
filo = r"C:\MyProject\Adaptive_Filtered_Single_Channel_Interferogram.pix"
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
Generate a filtered, multi-channel interferogram using a seven-by-seven adaptive Lee filter. The output file will contain four interferograms. Each output layer (n) is defined by the interferogram between the reference layer [DBIC_REF(n)] and the dependent layer [DBIC(n)].
from pci.insraw2 import insraw2
filref = 'Multi_Pol_Reference_File.pix'
dbic_ref = [1,2,3,4]
fili = 'Multi_Pol_Resampled_Dependent_File.pix'
dbic = [1,2,3,4]
flsz = [7]
filo = 'Adaptive_Filtered_Multi_Channel_Interferogram.pix'
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
from pci.insraw2 import insraw2
filref = r"C:\MyProject\Multi_Pol_Reference_File.pix"
dbic_ref = [1,2,3,4]
fili = r"C:\MyProject\Multi_Pol_Resampled_Dependent_File.pix"
dbic = [1,2,3,4]
flsz = [7]
filo = r"C:\MyProject\Adaptive_Filtered_Multi_Channel_Interferogram.pix"
insraw2(filref, dbic_ref, fili, dbic, flsz, filo)
| Back to top |
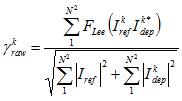


| Back to top |
© PCI Geomatics Enterprises, Inc.®, 2024. All rights reserved.